In nanopore sequencing, an electric field pulls ions in the water and strands of DNA through a minuscule protein hole or a hole in a solid-state membrane. Because the pore is not much wider than the DNA strand, when a strand passes through the amount of ionic current is altered. Each of the four nucleic acids in DNA—G, T, C, and A—whose sequence spells out the code for a living thing—can be identified by its distinct effect on the current.
But the current in question is very small, measured in picoamps. And the DNA passes through the pore at such a rapid clip that electronics have a difficult time distinguishing such a small signal in so short a time.
Where other nanopore systems operate at a rate of 10 to 100 kilohertz—not fast enough for DNA moving through the pore at roughly 1 million chemical units, or bases, per second—Xie says the Harvard version should in principle operate at a few gigahertz, far faster than the DNA moves, although they didn’t have the equipment to measure that.
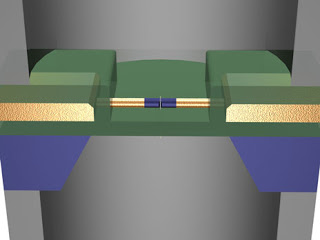
Image: Ping Xie and Charles M. Lieber/Harvard University DNA SLIPSTREAM: Ionic current pushes DNA through a nanometer-scale pore and past a nanowire transistor [blue center]. The transistor amplifies the change in current allowing the DNA’s sequence to be read
Nature Nanotechnology – Local electrical potential detection of DNA by nanowire–nanopore sensors
An individual genome is a sequence of about 3 billion nucleic acids, so even at high speeds, sequencing with a single nanopore would take too long. Practical sequencers would have to consist of multiple nanopores. However, in previous nanopore sequencer designs, electrical cross talk would occur between adjacent nanopores unless each one was secluded in its own chamber of solution. The Harvard version relies on the highly localized voltage, which is concentrated within just 30 to 50 nanometers of the opening, preventing cross talk with other nanopores as long as they are at least a few micrometers apart. Xie says the design should allow many nanopores to be grouped together on a single chip with shared solution chamber
Nanopores could potentially be used to perform single-molecule DNA sequencing at low cost and with high throughput. Although single base resolution and differentiation have been demonstrated with nanopores using ionic current measurements direct sequencing has not been achieved because of the difficulties in recording very small (~pA) ionic currents at a bandwidth consistent with fast translocation speeds. Here, we show that solid-state nanopores can be combined with silicon nanowire field-effect transistors to create sensors in which detection is localized and self-aligned at the nanopore. Well-defined field-effect transistor signals associated with DNA translocation are recorded when an ionic strength gradient is imposed across the nanopores. Measurements and modelling show that field-effect transistor signals are generated by highly localized changes in the electrical potential during DNA translocation, and that nanowire–nanopore sensors could enable large-scale integration with a high intrinsic bandwidth.
20 pages of supplemental material
If you liked this article, please give it a quick review on ycombinator or StumbleUpon. Thanks

Brian Wang is a Futurist Thought Leader and a popular Science blogger with 1 million readers per month. His blog Nextbigfuture.com is ranked #1 Science News Blog. It covers many disruptive technology and trends including Space, Robotics, Artificial Intelligence, Medicine, Anti-aging Biotechnology, and Nanotechnology.
Known for identifying cutting edge technologies, he is currently a Co-Founder of a startup and fundraiser for high potential early-stage companies. He is the Head of Research for Allocations for deep technology investments and an Angel Investor at Space Angels.
A frequent speaker at corporations, he has been a TEDx speaker, a Singularity University speaker and guest at numerous interviews for radio and podcasts. He is open to public speaking and advising engagements.