Synthetic biology researchers at Northwestern University, working with partners at Harvard Medical School, have for the first time synthesized ribosomes — cell structures responsible for generating all proteins and enzymes in our bodies — from scratch in a test tube.
Customized ribosomes could lead to new antibiotics and new proteins.
Others have previously tried to synthesize ribosomes from their constituent parts, but the efforts have yielded poorly functional ribosomes under conditions that do not replicate the environment of a living cell. In addition, attempts to combine ribosome synthesis and assembly in a single process have failed for decades.
* An integrated synthesis, assembly, and translation technology (termed iSAT) was developed to construct ribosomes in vitro.
* iSAT mimics co-transcription of rRNA and ribosome assembly as it occurs in vivo.
* iSAT makes possible the in vitro construction of modified ribosomes.
* iSAT is expected to aid studies of ribosome assembly and open new avenues for making ribosomes with altered capabilities
Michael C. Jewett, a synthetic biologist at Northwestern, George M. Church, a geneticist at Harvard Medical School, and colleagues recently took another approach: they mimicked the natural synthesis of a ribosome, allowing natural enzymes of a cell to help facilitate the man-made construction.
The technology could lead to the discovery of new antibiotics targeting ribosome assembly; an advanced understanding of how ribosomes form and function; and the creation of tailor-made ribosomes to produce new proteins with exotic functions that would be difficult, if not impossible, to make in living organisms.
“We can mimic nature and create ribosomes the way nature has evolved to do it, where all the processes are co-activated at the same time,” said Jewett, who led the research along with Church. “Our approach is a one-pot synthesis scheme in which we toss genes encoding ribosomal RNA, natural ribosomal proteins, and additional enzymes of an E. coli cell together in a test tube, and this leads to the construction of a ribosome.”
Jewett is an assistant professor of chemical and biological engineering at Northwestern’s McCormick School of Engineering and Applied Science.
The in vitro construction of ribosomes, as demonstrated in this study, is of great interest to the synthetic biology field, which seeks to transform the ability to engineer new or novel life forms and biocatalytic ensembles for useful purposes.
The findings of the four-year research project were published June 25 in the journal Molecular Systems Biology.
Comprising 57 parts — three strands of ribonucleic acid (RNA) and 54 proteins — ribosomes carry out the translation of messenger RNA into proteins, a core process of the cell. The thousands of proteins per cell, in turn, carry out a vast array of functions, from digestion to the creation of antibodies. Cells require ribosomes to live.
Jewett likens a ribosome to a chef. The ribosome takes the recipe, encoded in DNA, and makes the meal, or a protein. “We want to make brand new chefs, or ribosomes,” Jewett said. “Then we can alter ribosomes to do new things for us.”
“The ability to make ribosomes in vitro in a process that mimics the way biology does it opens new avenues for the study of ribosome synthesis and assembly, enabling us to better understand and possibly control the translation process,” he said. “Our technology also may enable us in the future to rapidly engineer modified ribosomes with new behaviors and functions, a potentially significant advance for the synthetic biology field.”
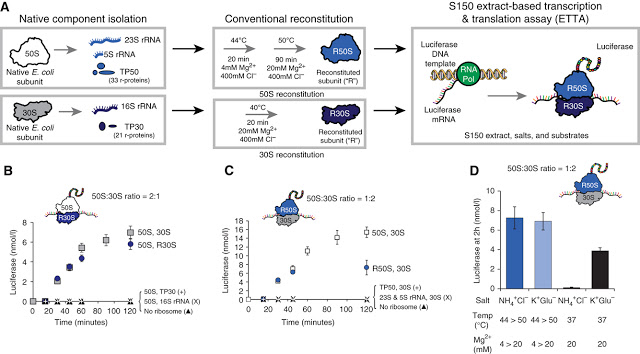
The synthesis process developed by Jewett and Church — termed “integrated synthesis, assembly and translation” (iSAT) technology — mimics nature by enabling ribosome synthesis, assembly and function in a single reaction and in the same compartment.
Working with E. coli cells, the researchers combined natural ribosomal proteins with synthetically made ribosomal RNA, which self-assembled in vitro to create semi-synthetic, functional ribosomes.
They confirmed the ribosomes were active by assessing their ability to carry out translation of luciferase, the protein responsible for allowing a firefly to glow. The researchers then showed the ability of iSAT to make a modified ribosome with a point mutation that mediates resistance to the antibiotic clindamycin.
The researchers next want to synthesize all 57 ribosome parts, including the 54 proteins.
“I’m really excited about where we are,” Jewett said. “This study is an important step along the way to synthesizing a complete ribosome. We will continue to push this work forward.”
ABSTRACT – Purely in vitro ribosome synthesis could provide a critical step towards unraveling the systems biology of ribosome biogenesis, constructing minimal cells from defined components, and engineering ribosomes with new functions. Here, as an initial step towards this goal, we report a method for constructing Escherichia coli ribosomes in crude S150 E. coli extracts. While conventional methods for E. coli ribosome reconstitution are non-physiological, our approach attempts to mimic chemical conditions in the cytoplasm, thus permitting several biological processes to occur simultaneously. Specifically, our integrated synthesis, assembly, and translation (iSAT) technology enables one-step co-activation of rRNA transcription, assembly of transcribed rRNA with native ribosomal proteins into functional ribosomes, and synthesis of active protein by these ribosomes in the same compartment. We show that iSAT makes possible the in vitro construction of modified ribosomes by introducing a 23S rRNA mutation that mediates resistance against clindamycin. We anticipate that iSAT will aid studies of ribosome assembly and open new avenues for making ribosomes with altered properties.
If you liked this article, please give it a quick review on ycombinator or StumbleUpon. Thanks

Brian Wang is a Futurist Thought Leader and a popular Science blogger with 1 million readers per month. His blog Nextbigfuture.com is ranked #1 Science News Blog. It covers many disruptive technology and trends including Space, Robotics, Artificial Intelligence, Medicine, Anti-aging Biotechnology, and Nanotechnology.
Known for identifying cutting edge technologies, he is currently a Co-Founder of a startup and fundraiser for high potential early-stage companies. He is the Head of Research for Allocations for deep technology investments and an Angel Investor at Space Angels.
A frequent speaker at corporations, he has been a TEDx speaker, a Singularity University speaker and guest at numerous interviews for radio and podcasts. He is open to public speaking and advising engagements.