Researchers demonstrated that by altering the length of Cas9-associated guide RNA (gRNA) we were able to control Cas9 nuclease activity and simultaneously perform genome editing and transcriptional regulation with a single Cas9 protein. We exploited these principles to engineer mammalian synthetic circuits with combined transcriptional regulation and kill functions governed by a single multifunctional Cas9 protein.
Initially, the CRISPR gene editing system took advantage of the Cas9 enzyme, a naturally occurring protein in the immune system of certain bacteria, which acts like a pair of molecular scissors to precisely cut or edit specific sections of DNA. However, scientists have recently begun to manipulate CRISPR-Cas9 variants as gene regulation tools, in order to reversibly turn genes on or off.
Both the gene editing and gene regulation processes start with the same step, which is the recruitment of Cas9 to the genes of interest by a matching sequence comprised of guide RNA, which aids the Cas9 attachment onto the DNA sequence. Yet, until now, investigators required the use of two variants of the Cas9 protein to perform either the gene editing or the gene regulation steps.
Currently, researchers from Harvard University and The Massachusetts Institute of Technology have developed a new approach that will allow researchers to achieve both tasks using a single Cas9 isoform. The Harvard/MIT team found that the length of the guide RNA sequence played a critical role in determining the fate of Cas9—whether it would solely bind to DNA or if it would excise it as well.
“We decided to systematically test why it was that truncating the guides too much caused Cas9 to no longer cut the intended genomic site,” stated Alejandro Chavez, Ph.D., postdoctoral fellow at Harvard’s Wyss Institute for Biologically Inspired Engineering.
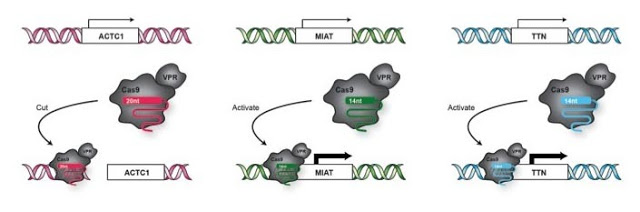
Cells were transfected with a 20-nt guide directed toward ACTC1, 14-nt guides directed toward MIAT and TTN and either Cas9-VPR or Cas9. This picture represents the expected behavior of Cas9-VPR. The ACTC1 locus should be cut, while transcription occurs for the genes MIAT and ACTC1.
Activation and cutting of a transcriptional reporter using gRNAs with progressively shorter 5ʹ end lengths.
Simplified schematics of circuit
Nature Methods – Cas9 gRNA engineering for genome editing, activation and repression
The researchers found that shorter guide RNAs prevented Cas9 from cleaving the target gene, but not from efficiently binding to the target. This finding could open up the possibility for scientists to attach gene regulation proteins to Cas9 for delivery to specific genes.
“By using our uncovered guide RNA principles, we can now for the first time toggle a single protein to gain direct control over both, gene sequences and gene expression, and turn almost any DNA sequence into a regulatory sequence to further bend the cell to our will,” explained co-senior author George Church. Ph.D., professor of health sciences and technology at Harvard and MIT. “We envision future uses for the technology that can help decipher the tangled web of interactions underlying for example cancer drug resistance and stem cell differentiation, or design advanced synthetic gene circuitries.”
“This new functionality will improve our ability to decipher the complex relationships between interdependent genes responsible for many diseases,” noted Marcelle Tuttle, Ph.D., research fellow at the Wyss Institute and co-author on the current study.
The investigators were excited by their findings and believe that this new method could also be used in large-scale metabolic production of chemicals and fuels using genetically engineered bacteria, while simultaneously safeguarding the microbial biofactories from infection by foreign pathogens.
SOURCES – Nature Methods, Genengnews

Brian Wang is a Futurist Thought Leader and a popular Science blogger with 1 million readers per month. His blog Nextbigfuture.com is ranked #1 Science News Blog. It covers many disruptive technology and trends including Space, Robotics, Artificial Intelligence, Medicine, Anti-aging Biotechnology, and Nanotechnology.
Known for identifying cutting edge technologies, he is currently a Co-Founder of a startup and fundraiser for high potential early-stage companies. He is the Head of Research for Allocations for deep technology investments and an Angel Investor at Space Angels.
A frequent speaker at corporations, he has been a TEDx speaker, a Singularity University speaker and guest at numerous interviews for radio and podcasts. He is open to public speaking and advising engagements.