The cutting protein Cas9 can be replaced by a different protein, Cpf1, which Feng Zhang, a researcher at the Broad Institute of MIT and Harvard says will also work as a versatile editing tool. The background for the Broad announcement is a bruising patent fight with the University of California, Berkeley, over who invented the first CRISPR editing tools, in particular Cas9.
Zheng was the scientist who first harnessed the revolutionary CRISPR-Cas9 system for mammalian genome editing has now identified a different CRISPR system with the potential for even simpler and more precise genome engineering.
Zhang and his collaborators searched through hundreds of CRISPR systems in different types of bacteria, searching for enzymes with useful properties that could be engineered for use in human cells. Two promising candidates were the Cpf1 enzymes from bacterial species Acidaminococcus and Lachnospiraceae, which Zhang and his colleagues then showed can target genomic loci in human cells.
The new system, because it has a different cutting protein, could offer a way around the legal quagmire. “The greatest value may be more in terms of the patent landscape than a scientific advancement,” says Dan Voytas, a genome-editing researcher at the University of Minnesota.
The stakes are high as startups race to develop gene editing as a basis for possible medical treatments. Editas Medicine, which is connected with Feng’s lab, raised an additional $120 million in August. Intellia, a competitor connected to the Berkeley team, raised $70 million this month.
CRISPR is based on a natural system some bacteria use to defend against viruses by shredding their invading genes. In the laboratory, it’s been adapted as a tool that consists of two key components: a short stretch of RNA that lines up with a specific gene, and then a cutting protein that moves in to snip the gene open.
Journal Cell – Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System
Highlights
•CRISPR-Cpf1 is a class 2 CRISPR system
•Cpf1 is a CRISPR-associated two-component RNA-programmable DNA nuclease
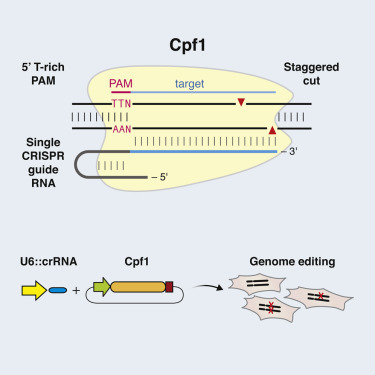
•Targeted DNA is cleaved as a 5-nt staggered cut distal to a 5′ T-rich PAM
•Two Cpf1 orthologs exhibit robust nuclease activity in human cells
Summary
The microbial adaptive immune system CRISPR mediates defense against foreign genetic elements through two classes of RNA-guided nuclease effectors. Class 1 effectors utilize multi-protein complexes, whereas class 2 effectors rely on single-component effector proteins such as the well-characterized Cas9. Here, we report characterization of Cpf1, a putative class 2 CRISPR effector. We demonstrate that Cpf1 mediates robust DNA interference with features distinct from Cas9. Cpf1 is a single RNA-guided endonuclease lacking tracrRNA, and it utilizes a T-rich protospacer-adjacent motif. Moreover, Cpf1 cleaves DNA via a staggered DNA double-stranded break. Out of 16 Cpf1-family proteins, we identified two candidate enzymes from Acidominococcus and Lachnospiraceae, with efficient genome-editing activity in human cells. Identifying this mechanism of interference broadens our understanding of CRISPR-Cas systems and advances their genome editing applications.
he newly described Cpf1 system differs in several important ways from the previously described Cas9, with significant implications for research and therapeutics, as well as for business and intellectual property:
1. In its natural form, the DNA-cutting enzyme Cas9 forms a complex with two small RNAs, both of which are required for the cutting activity. The Cpf1 system is simpler in that it requires only a single RNA. The Cpf1 enzyme is also smaller than the standard SpCas9, making it easier to deliver into cells and tissues.
2. Significantly, Cpf1 cuts DNA in a different manner than Cas9. When the Cas9 complex cuts DNA, it cuts both strands at the same place, leaving ‘blunt ends’ that often undergo mutations as they are rejoined. With the Cpf1 complex the cuts in the two strands are offset, leaving short overhangs on the exposed ends. This is expected to help with precise insertion, allowing researchers to integrate a piece of DNA more efficiently and accurately.
3. Cpf1 cuts far away from the recognition site, meaning that even if the targeted gene becomes mutated at the cut site, it can likely still be re-cut, allowing multiple opportunities for correct editing to occur.
4. The Cpf1 system provides new flexibility in choosing target sites. Like Cas9, the Cpf1 complex must first attach to a short sequence known as a PAM, and targets must be chosen that are adjacent to naturally occurring PAM sequences. The Cpf1 complex recognizes very different PAM sequences from those of Cas9. This could be an advantage in targeting some genomes, such as in the malaria parasite as well as in humans.
The new system, because it has a different cutting protein, could offer a way around the legal quagmire. “The greatest value may be more in terms of the patent landscape than a scientific advancement,” says Dan Voytas, a genome-editing researcher at the University of Minnesota.
The stakes are high as startups race to develop gene editing as a basis for possible medical treatments. Editas Medicine, which is connected with Feng’s lab, raised an additional $120 million in August. Intellia, a competitor connected to the Berkeley team, raised $70 million this month.
CRISPR is based on a natural system some bacteria use to defend against viruses by shredding their invading genes. In the laboratory, it’s been adapted as a tool that consists of two key components: a short stretch of RNA that lines up with a specific gene, and then a cutting protein that moves in to snip the gene open.
SOURCES – MIT Technology Review, Journal Cell, Broad Institute of MIT and Harvard

Brian Wang is a Futurist Thought Leader and a popular Science blogger with 1 million readers per month. His blog Nextbigfuture.com is ranked #1 Science News Blog. It covers many disruptive technology and trends including Space, Robotics, Artificial Intelligence, Medicine, Anti-aging Biotechnology, and Nanotechnology.
Known for identifying cutting edge technologies, he is currently a Co-Founder of a startup and fundraiser for high potential early-stage companies. He is the Head of Research for Allocations for deep technology investments and an Angel Investor at Space Angels.
A frequent speaker at corporations, he has been a TEDx speaker, a Singularity University speaker and guest at numerous interviews for radio and podcasts. He is open to public speaking and advising engagements.