There is a new gene-editing tool called Retron Library Recombineering (RLR) that makes this task easier that has been developed by George Church and his team. RLR generates up to millions of mutations simultaneously, and “barcodes” mutant cells so that the entire pool can be screened at once, enabling massive amounts of data to be easily generated and analyzed. The achievement, which has been accomplished in bacterial cells.
RLR enabled us to do something that’s impossible to do with CRISPR: we randomly chopped up a bacterial genome, turned those genetic fragments into single-stranded DNA in situ, and used them to screen millions of sequences simultaneously,” said co-first author Max Schubert, Ph.D., a postdoc in the lab of Wyss Core Faculty member George Church, Ph.D. “RLR is a simpler, more flexible gene editing tool that can be used for highly multiplexed experiments, which eliminates the toxicity often observed with CRISPR and improves researchers’ ability to explore mutations at the genome level.
Retrons’ existence has been known for decades, but the function of the ssDNA they produce flummoxed scientists from the 1980s until June 2020, when a team finally figured out that retron ssDNA detects whether a virus has infected the cell, forming part of the bacterial immune system.
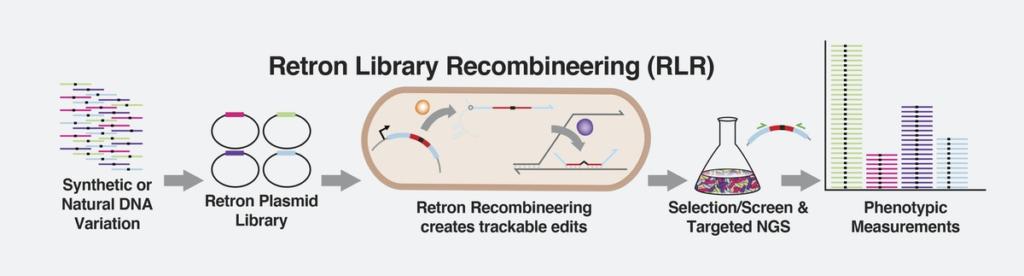
Retrons enable the rapid production and screening of millions of trackable DNA variations and their effects on bacteria simultaneously. Credit: Max Schubert / Wyss Institute at Harvard University
Retrons: from enigma to engineering tool
Retrons are segments of bacterial DNA that undergo reverse transcription to produce fragments of single-stranded DNA (ssDNA). Retrons’ existence has been known for decades, but the function of the ssDNA they produce flummoxed scientists from the 1980s until June 2020, when a team finally figured out that retron ssDNA detects whether a virus has infected the cell, forming part of the bacterial immune system.
While retrons were originally seen as simply a mysterious quirk of bacteria, researchers have become more interested in them over the last few years because they, like CRISPR, could be used for precise and flexible gene editing in bacteria, yeast, and even human cells.
“For a long time, CRISPR was just considered a weird thing that bacteria did, and figuring out how to harness it for genome engineering changed the world. Retrons are another bacterial innovation that might also provide some important advances,” said Schubert. His interest in retrons was piqued several years ago because of their ability to produce ssDNA in bacteria – an attractive feature for use in a gene editing process called oligonucleotide recombineering.
To see if they could actually use retrons to achieve efficient recombineering with retrons, Schubert and his colleagues first created circular plasmids of bacterial DNA that contained antibiotic resistance genes placed within retron sequences, as well as an SSAP gene to enable integration of the retron sequence into the bacterial genome. They inserted these retron plasmids into E. coli bacteria to see if the genes were successfully integrated into their genomes after 20 generations of cell replication. Initially, less than 0.1% of E. coli bearing the retron recombineering system incorporated the desired mutation.
To improve this disappointing initial performance, the team made several genetic tweaks to the bacteria. First, they inactivated the cells’ natural mismatch repair machinery, which corrects DNA replication errors and could therefore be “fixing” the desired mutations before they were able to be passed on to the next generation. They also inactivated two bacterial genes that code for exonucleases – enzymes that destroy free-floating ssDNA. These changes dramatically increased the proportion of bacteria that incorporated the retron sequence, to more than 90% of the population.
“Being able to analyze pooled, barcoded mutant libraries with RLR enables millions of experiments to be performed simultaneously, allowing us to observe the effects of mutations across the genome, as well as how those mutations might interact with each other,” said senior author George Church, who leads the Wyss Institute’s Synthetic Biology Focus Area and is also a Professor of Genetics at HMS. “This work helps establish a road map toward using RLR in other genetic systems, which opens up many exciting possibilities for future genetic research.”
Another feature that distinguishes RLR from CRISPR is that the proportion of bacteria that successfully integrate a desired mutation into their genome increases over time as the bacteria replicate, whereas CRISPR’s “one shot” method tends to either succeed or fail on the first try. RLR could potentially be combined with CRISPR to improve its editing performance, or could be used as an alternative in the many systems in which CRISPR is toxic.
More work remains to be done on RLR to improve and standardize editing rate, but excitement is growing about this new tool. RLR’s simple, streamlined nature could enable the study of how multiple mutations interact with each other, and the generation of a large number of data points that could enable the use of machine learning to predict further mutational effects.
PNAS – High-throughput functional variant screens via in vivo production of single-stranded DNA
Significance
We report a methodology for the pooled construction of mutants bearing precise genomic sequence variations and multiplex phenotypic characterization of these mutants using next-generation sequencing (NGS). Unlike existing techniques depending on CRISPR-Cas–directed genomic breaks for genome editing, this strategy instead uses single-stranded DNA produced by a retron element for recombineering. This enables libraries of millions of elements to be constructed and offers relaxed design constraints which permit natural DNA or random variation to be used as inputs.
Abstract
Creating and characterizing individual genetic variants remains limited in scale, compared to the tremendous variation both existing in nature and envisioned by genome engineers. Here we introduce retron library recombineering (RLR), a methodology for high-throughput functional screens that surpasses the scale and specificity of CRISPR-Cas methods. We use the targeted reverse-transcription activity of retrons to produce single-stranded DNA (ssDNA) in vivo, incorporating edits at >90% efficiency and enabling multiplexed applications. RLR simultaneously introduces many genomic variants, producing pooled and barcoded variant libraries addressable by targeted deep sequencing. We use RLR for pooled phenotyping of synthesized antibiotic resistance alleles, demonstrating quantitative measurement of relative growth rates. We also perform RLR using the sheared genomic DNA of an evolved bacterium, experimentally querying millions of sequences for causal variants, demonstrating that RLR is uniquely suited to utilize large pools of natural variation. Using ssDNA produced in vivo for pooled experiments presents avenues for exploring variation across the genome.
SOURCES – PNAS, Wyss Institute at Harvard
Written by Brian Wang, Nextbigfuture.com

Brian Wang is a Futurist Thought Leader and a popular Science blogger with 1 million readers per month. His blog Nextbigfuture.com is ranked #1 Science News Blog. It covers many disruptive technology and trends including Space, Robotics, Artificial Intelligence, Medicine, Anti-aging Biotechnology, and Nanotechnology.
Known for identifying cutting edge technologies, he is currently a Co-Founder of a startup and fundraiser for high potential early-stage companies. He is the Head of Research for Allocations for deep technology investments and an Angel Investor at Space Angels.
A frequent speaker at corporations, he has been a TEDx speaker, a Singularity University speaker and guest at numerous interviews for radio and podcasts. He is open to public speaking and advising engagements.


Can you point to any time in history when medicine was not focused mostly on improving the damaged?
If for no other reason than it's much easier to do.
This scares me
Improving the damaged rather than optimizing the healthy is a recent and disturbing cultural phenomena
I don't know. You spend a fortune to get vitamin generating gut flora installed, then you eat a dodgy kebab next month, spend a couple of days on the toilet, a week on antibiotics, and now you're back to normal.
more interest in designer kids than GMOs for some reason in UK/EU.
Is One suggesting designer Kids/ Yuppies/ Fogies?
geez, maybe more like a research grenade. Hopefully such friendly fire 'fragments' provide value.
"generates up to millions of mutations simultaneously"
This! This is why we get Swamp Things and Hulks, people!
Seriously, if George Church is pleased with it then so am I.
This couldn't be used as part of a biological warefare campaign, right?
I would think if you wanted to add Vitamin C synthesis, you'd do the engineering on our gut flora. You could do that for most of the vitamins, indeed we already get some of our vitamins that way.
Of course, this technique would be great for something like that.
The Synthetic Biology guys are going to be very important here. They are the ones developing the tools to manage the Objects (genetic sequences controlling protein synthesis) and the models that can predict how they will interact when compiled into a system (organism). Using techniques they stole from integrated circuit designers they can now design whole new genomes with extraordinary capabilities. First however, and this is where RLR comes in, they need to study how the genetic sequences they use actually DO function in an organic framework – only then can they confident that when it is used in a new design it will perform as desired.
I almost pity the bright young minds in University today – the plethora of choice they face in choosing what technology to specialize in is truly daunting – Synthetic Biology, Rocket Engineering, Robotics, AI, Nanotech….wow.
That last, as I read it. Sounds totally worthless for in vivo genetic engineering, though might be handy for in vitro.
There are a lot of genetic therapies where you wouldn't need to hit every cell, just introduce some cells with the right genes. Curing diabetes, for instance. In those cases, sample some tissue, make the edit, select for the ones that came out right, and reintroduce them.
Unfortunately, there are also a lot of genetic therapies where you'd need to fix cells in place, and this doesn't seem terribly handy for that.
I'm a little unclear on this. They have a method of making many simultaneous precise edits in the same cell, a single or few precise edits in many cells at once, or are they just dumping a bunch of many different potential editing sequences in a pool of cells and running a screen to see what survivors emerge?
Yeah… what could possibly go wrong?
AI? He prefers to be called Zuckerberg these days.
Well, as the article states, that is for the AI to determine.
While CRISPR is a gun, RLR is a machinegun, allowing many, many changes at once over a group of targeted cells.
This can circumvent the problem of wrong insertion errors that don't work or are fatal to the cells because they are too small, or those that don't work because they need several edits and the cells start dividing before they are ready, allowing more complex edits requiring a lot of changes at once, like most useful mutations tend to be.
This one looks like an actual quick genetic editing tool for new organisms, not requiring several generations to see results.
And of course, a genetic therapy tool over tissues and organs.
SOmething else to add to vaccines!
Though what is simple enough for modern tech to actually implement, that you would want to add to all human DNA, I don't know. Vitamin C synthesis? Lactose tolerance?? An inactive stretch of Pig DNA just to piss off 1 billion Muslims??? (Though I suspect that Swine flu would already have DNA in it that was originally in pigs…)